There are more viruses than cells on this planet. Understanding viruses is important because they cause many severe diseases, but they also provide good models for understanding the structure and behavior of proteins, nucleic acids and membranes. Viruses are a heterogeneous group and span in size from over 500 nm down to less than 20 nm. Some are round, some are fibrous and some have a complex composition. We therefore need to employ different techniques to study them.
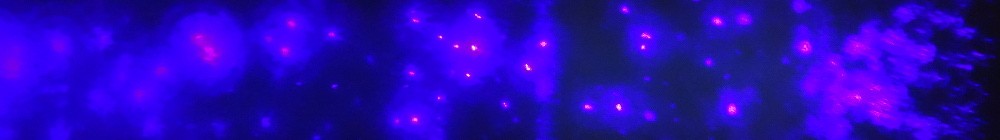
Virus imaging has traditionally relied on X-ray crystallography, but the field has in the last few years been expanding in the direction of single-particle methods. Our efforts in developing X-FEL based imaging is one such initiative and could provide possibilities to study non-symmetric features, such as the virus particle interior structures. As the first group in the world we have been imaging single particles of viruses by injection into vacuum and illumination by X-rays from the free-electron laser at the Linear Coherent Light Source (LCLS) [1]. Challenges to this technique are sample delivery and software development for data processing, such as phasing and reconstruction.
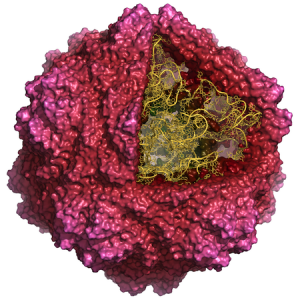
Some aspects of viruses are very difficult or impossible to study experimentally and therefore we also employ models and simulations to do experiments in the computer. Based on classical Newtonian physics one can build up an all-atom model of an entire virus particle and simulate it over microsecond time-scale to predict behavior regarding disassembly [2] and genome packing [3].
Publications:
- Seibert, Ekeberg, Maia, et al., Nature 470: 78–81 (2011). Read online
- Larsson, et al., PLoS Comput. Biol. 8: e1002502 (2012). Read online
- Larsson, et al., J. of Chem. Theory and Comput. 8: 2474–2483 (2012). Read online