Biology is heterogeneous: its building blocks undergo changes themselves. Variations in form, function with a resulting change in behavior exist everywhere; in proteins, in cells, up to entire organisms. This heterogeneity can be overwhelming, but to understand the organization of life, we need to characterise its heterogeneity.
Structural heterogeneity of proteins
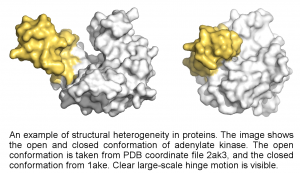
One part of this project is the structural heterogeneity of proteins. Proteins often do not have one unique structure, but interchange between several conformational states. The differences between these states can be large, such as the motion of entire domains (see figure), or small such as changes around the active site. In either case, these changes are essential for the function of the protein.
In an XFEL experiment, the ultra-short X-ray pulse captures the protein in a single conformation unlike many imaging techniques where the exposure time is much slower than the movements within the sample. By imaging many individual proteins, we can sample many of the proteins conformation, ultimately allowing us to recover the entire conformational space of the protein.
There is yet no data that allows this kind of study, we are instead focusing on developing the necessary algorithms and software using simulated data. We do however expect for the first experimental results within the next few years.
Heterogeneity in organelles
Our carboxysome project is dealing with sample heterogeneity by classifying diffraction patterns and reconstructing projection images in an automised processing pipeline. The individual reconstructions have a resolution of 22 nm, to date the highest reported resolution of a single biological particle imaged with an XFEL. Moreover, the results show that the injection process does not alter the size distribution of the particles.
Imaging single live cells
Another of our projects studied single living cells. It order to avoid the overwhelming amount of heterogeneity, it examined single 2D reconstructions at a time. Predictions showed that 2D reconstructions could reach sub-nanometre resolution on micron-sized cells. We developed a container-free injection method that is capable of introducing single living cells into the X-ray pulse. This method minimises the background scattering, and is suitable for high-throughput studies. Currently, our reconstructions reach 80 nm resolution, but are mainly limited by the dynamic range of the detectors. Most intriguingly, we are already recording diffraction signal up to 4 nm resolution.
Outlook
Life is more than a sum of its parts. However, currently structural biology is limited to studying exactly the parts. In order to understand the whole, i.e. cells, we would need a technique that can examine the entire cell, without losing the resolution required to understand the parts. We believe CXDI could be such a technique. Our future goal is to obtain 3D information of living cells at sub-nanometer resolution. Small changes on the level of individual proteins would be visible, as well as the resulting change in the global network. This knowledge will have tremendous effects on our fundamental understanding of cellular life.